My name is Matthew Moulton and I am a rising senior attending The Cooper Union for the Advancement of Science Art. After I graduate I plan to work as a chemical engineer. I applied to the NYU MRSEC REU program to gain experience and to explore a different scientific field. As part of the program, I am working as a research assistant in the Montclare Lab located in New York University Tandon School of Engineering.
This laboratory focuses on protein engineering. Essentially, the researchers create proteins with a desired outcome. For the summer, I am working with the protein Q. Q’s structure is best described as a bundle of coils. Previous research has shown that at high enough concentrations Q forms fibers that cross-link to form a hydrogel. A hydrogel is a network of polymer chains that are hydrophilic. Hydrogels are used for drug delivery and tissue engineering but most hydrogels are made from synthetic polymers. Hydrogels made from proteins like Q are more bio compatible. My job is to determine the range of conditions that this gel can form under.
Q Protein
In order to produce Q ,researchers use bacteria as the factories to produce protein. Here are the steps:
- The first step in this process is transformation. In this step a DNA (also known as plasmid, shown below in red) that encodes the Q protein is inserted into the bacteria host or factory. For our project we use a heat shock protocol. When the bacteria are exposed to high temperatures, their cell walls become permeable, allowing for the plasmid to get into the cell. The bacteria are transferred onto plates with nutrients that contain antibiotics. Normally, antibiotics kill bacteria. The bacteria that we use are resistant to antibiotics because the DNA plasmid contains a gene or set of genes that can breakdown antibiotics. This ensures that only cells containing the DNA grow on the plate.
- The plates are left to incubate overnight to allow the bacteria to reproduce. The following day colonies, which are small clusters of cells, are chosen and grown in a solution containing antibiotics. This solution, called media, contains the nutrients necessary for bacteria to survive and reproduce.
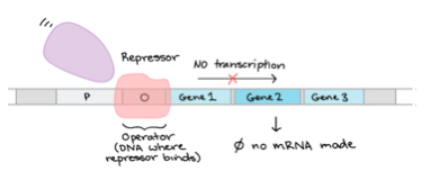
- This bacterial solution is used to initiate a larger volume of media and allowed to incubate until there are a sufficient amount of duplicated cells. Afterwards, a chemical, isopropyl β-D-1-thiogalactopyranoside, is added to the mixture to trigger the production of the Q protein. Isopropyl β-D-1-thiogalactopyranoside binds to a repressor protein (in pink below) on the DNA(shown below at the operator) and changes the repressor so it can no longer bind to the DNA. Once it comes off, another protein RNA polymerase (in purple) can take its place and begin interpreting the DNA so it can produce protein.
While this is just the first step of my experiments, I have learned a lot. I already knew how to perform transformation from a class I took at my university but I never learned how to produce protein. I was surprised at how many steps were involved. Protein production or expression is a process that takes several hours because once isopropyl β-D-1-thiogalactopyranoside is added to the solution, one has to wait for three hours for the cells to multiply.
For me, the most difficult part of protein expression was picking colonies. To pick a colony, a pipette tip is used to gently scrape a colony from the plate and place it in a tube. The first time I tried to pick a colony, I had trouble finding one because the colonies were so small so I accidentally poked through the plate!
I am still new and I make mistakes but I look forward to learning more about protein engineering. Do you have any advice to share as I learn more about protein engineers?
Let’s chat on Twitter:@matthewantonym
Matthew Anthony Moulton