By: Jason Chen
When I joined the Montclare lab for protein engineering and molecular design in the summer of 2020 (amidst a pandemic), I was eagerly expecting to be thrown into the wet lab world involving the prolific use of SDS-PAGE and the omnipresence of micropipette tips. Instead, I would find myself navigating a Unix shell often in the comfort of my bed (which was not great on my back, in retrospect). Scientific research is not all about wet lab work, as one might presume. In fact, computational work is one of the core driving forces of breakthroughs and advancements, especially in the field of protein engineering.
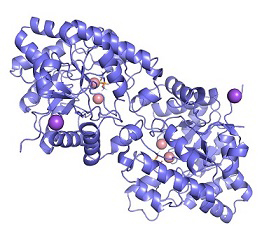
During my first few weeks, the learning curve was very steep. Although I was armed with three semesters of coding experience from my computer science courses, I was still mystified by the Unix interface and the modeling software called Rosetta. I perused through countless random Github and Stack Overflow web pages trying to figure out how to execute basic commands. I read and reread past papers about our protein construct (phosphotriesterase or PTE) to understand the meaning behind the work.
Most importantly, I received guidance and encouragement from my mentor, the patient and wise post-doctoral associate Farbod Mahmoudinobar. It also didn’t hurt to have worked with other undergrads on the project, Bonnie and Jakub, who sympathized with the struggles of undertaking computational work for the first time. We often connected through Zoom to troubleshoot difficulties and work together throughout the learning process.
Eventually, I grew more comfortable with the project. I gained a better understanding of the background of PTE as well as the overarching goals of our project. I was finally equipped with the necessary mental tools to computationally enhance the properties of PTE by altering its amino acids. Just as I was beginning to think that I had learned all I needed to know, I was faced with a whole new set of questions to answer. That’s the beauty of science.
Sure, I was now able to use the modeling software to modify our target protein, but this was just the tip of the iceberg. How should I interpret the output information from the new models? How could I use the knowledge from past design iterations to improve future designs? And the one that nagged at my brain most of all: why is a phenylalanine in this particular position and orientation more favorable than a histidine in that particular position? Finding the answers to these questions would require the convergence of these newly learned computational skills and my undergraduate chemistry background.
Learning to use a protein modeling software is like learning a new language. You first master the vocabulary, syntax, and how to put together sentences that can be understood by others. However, being able to functionalize and make meaning of tens of thousands of simulated results is akin to sifting for gold nuggets in a sandy riverbed. It’s a daunting task, but someone must do it to find the hidden treasures. And once the gold is found, it’s possible that the gold was actually just a shiny rock, a piece of fool’s gold, and you must start the sifting process all over again. Maybe you even need to look in a new riverbed.
Distilling the results of a modeling software such as Rosetta necessitates patience, an ability to recognize patterns, and a level of competence in the building blocks of proteins called amino acids. The multidisciplinary nature of the work requires both understanding of the software as well as the science behind the results. Similar to wet lab work, successful computational work is as much a testament to proficiency of technique as it is to the ingenuity and breadth of knowledge held by the researcher.
In my time here at the lab, I feel that I’ve only turned over one leaf in the forest that constitutes computational research. There is still so much to learn, and I am in awe of the broad scope of design applications that are possible through the Rosetta software and in powerful interactive interfaces such as the Unix shell. Although I’ve since started performing wet lab work with another mentor (the brilliant and wholesome PhD candidate, Jacob), I’m grateful to have been afforded the chance to step out of my comfort zone and explore an area of research I would not have been exposed to otherwise. Although computational work was often frustrating, like the many times I’d queue a Rosetta job for hours only for it to fail upon starting, the collaboration and the learning experience made it all worthwhile.